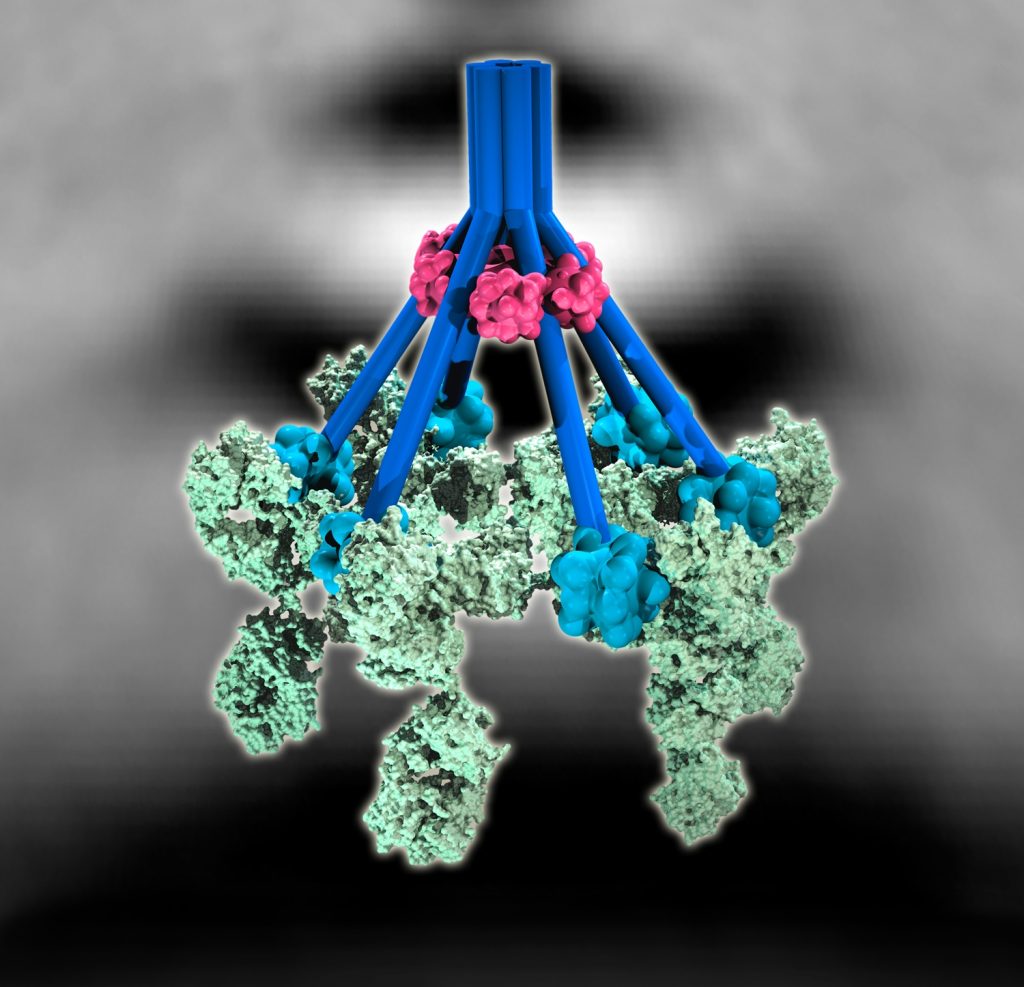
Researchers of Utrecht University, Intravacc and the National Institute for Public Health and the Environment used a new method to identify hitherto unknown peptide antigens. This type of antigen had long been searched for, as they may be the starting points for new vaccines and cancer immunotherapy. The results were published in Proceedings of the National Academy of Sciences (PNAS) on March 10, 2014.
“With this new method, we can identify more than twelve thousand peptide antigens, whereas before, we could only see the tip of the iceberg,” explains immunologist Dr Cécile van Els of the National Institute for Public Health and the Environment (RIVM). The new method, developed by the Biomolecular Mass Spectrometry and Proteomics group of Albert Heck, is not only much more sensitive, but also makes more routine research possible.
Complete picture
It is of vital importance for the development of vaccines and cancer immunotherapy to know which antigens alert the immune system. Up till now, it was not possible to get a complete picture of this process. In the best case, only a few hundreds of peptide antigens could be identified. But now, the researchers detecetd 12,000 of them, close to what is expected to be the maximum.
Cancer immunotherapy
The new method is also a very powerful technique for demonstrating the subtle differences between healthy cells and damaged cells, according to Geert Mommen of Intravacc. Of the twelve thousand peptide antigens, there are maybe a few dozen damaged ones that actually deviate from or are completely different from healthy cells. Researchers have found more proof that these deviating or unique peptide antigens are best equipped to prepare our immune system for the fight against cancer.
“Currently, a lot of research is being conducted into cancer immunotherapy,” adds Heck. “Science has even called this the break-through of the year 2013, but in actual fact, the therapy is still in its infancy. The good thing about immunotherapy is that it does not destroy or damage healthy cells. With our new method, researchers will obtain significantly more information about our immune system. We hope that this information will help them with the further development of cancer treatment.”
Read the full article:
Expanding the detectable HLA peptide repertoire using electron-transfer/higher-energy collision dissociation (EThcD)
Geert P. M. Mommen, Christian K. Frese, Hugo D. Meiring, Jacqueline van Gaans-van den Brink, Ad P. J. M. de Jong, Cécile A. C. M. van Els, Albert J. R. Heck
PNAS, Monday 10 March, doi 10.1073
This research is co-funded by the Netherlands Proteomics Centre (NPC), which performs high-quality research and transfer of knowledge in proteomics. The RIVM was part of this project as strategic research partner.