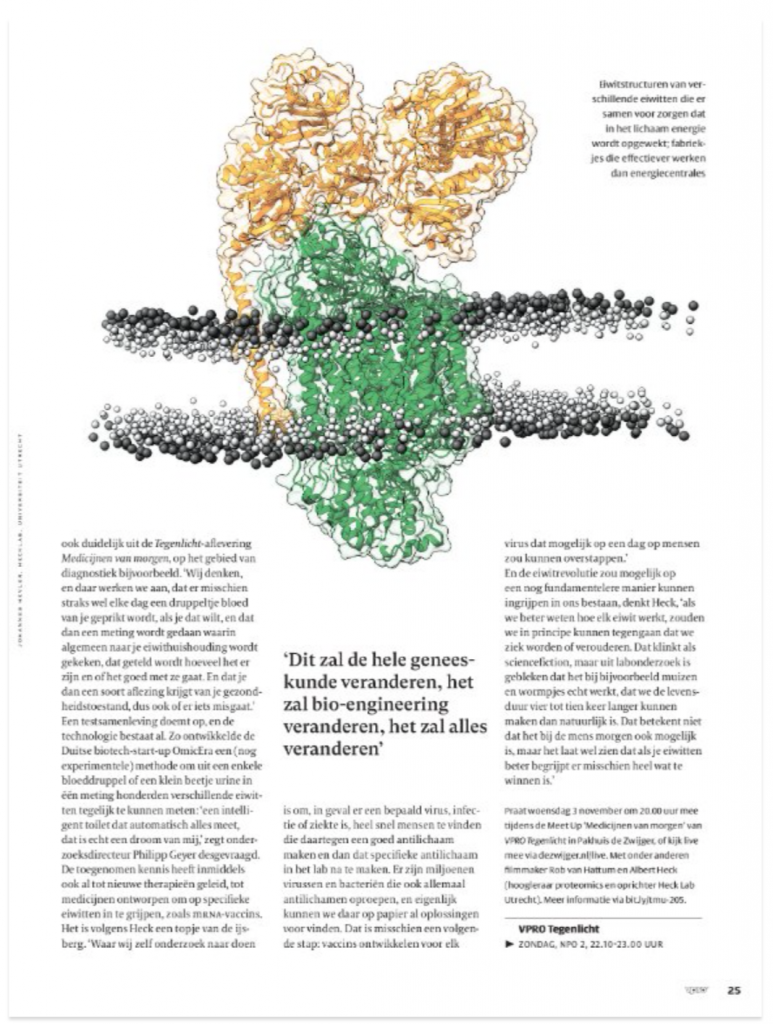
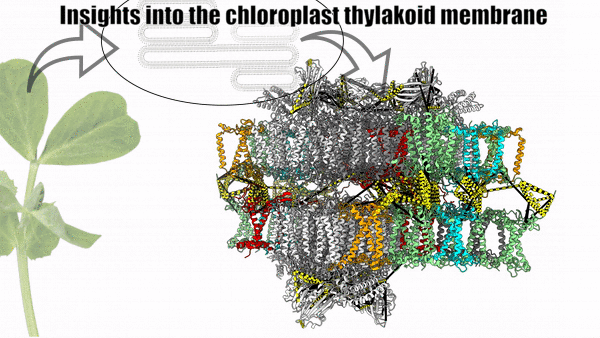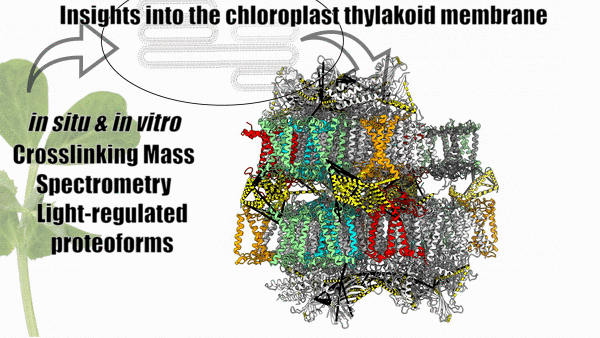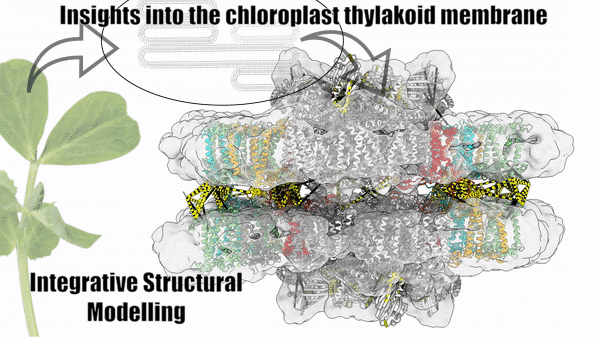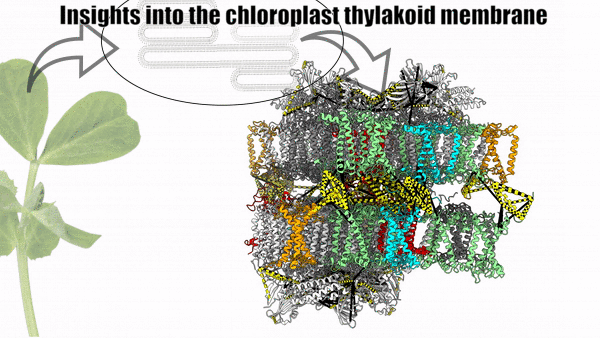
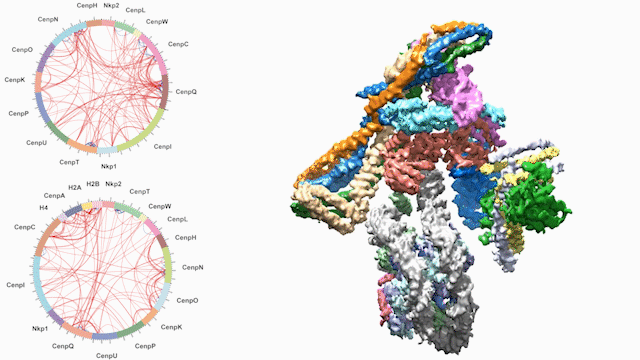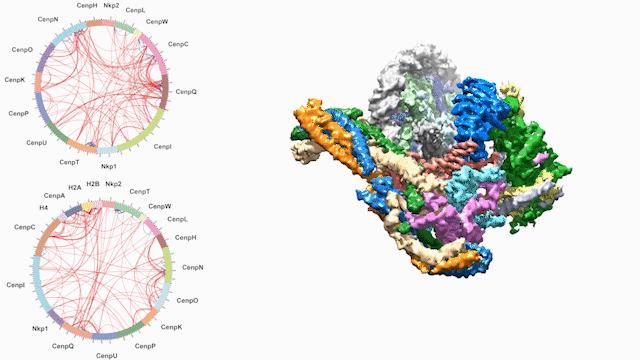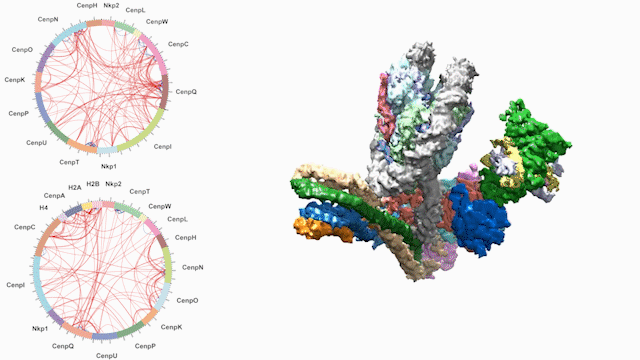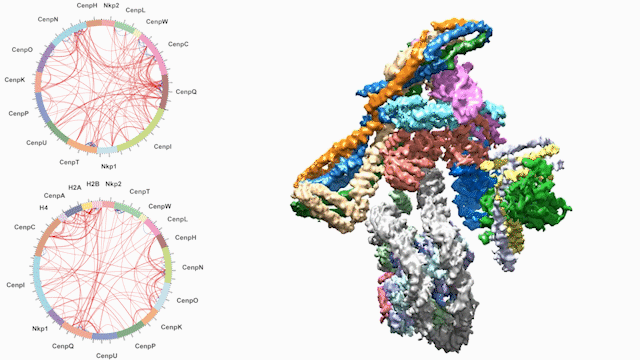
Dancing around
Using the method, the researchers discovered that the receptor continually changes shape in the absence of the EGF hormone. “We believe that the receptor in its ligand free state is continually switching from a ‘closed’ to an ‘open’ position and vice-versa”, says Paul van Bergen en Henegouwen, cellular biologist at Utrecht University and one of the authors of the article. “It so to speak dances around and continuously bows for its audience”.
New cancer drugs
In his lab, Van Bergen en Henegouwen develops antibody fragments, known as nanobodies, that can specifically bind to EGF receptors. These nanobodies can therefor be used to label tumour cells, which enables the specific destruction of such cells. “Since we now know more about the structure and dynamics of the EGF receptor, we can start designing nanobodies that specifically bind to the open, active form of the receptor. This can ultimately lead to new cancer drugs.”
Publication
‘EGFR Dynamics Change during Activation in Native Membranes as Revealed by NMR’
Cell, 10 november 2016, doi.org/10.1016/j.cell.2016.10.038
Mohammed Kaplan, Siddarth Narasimhan, Cecilia de Heus, Deni Mance,Sander van Doorn, Klaartje Houben, Dusan Popov-Celeketic, Reinier Damman, Eugene A. Katrukha, Purvi Jain, Willie J.C. Geerts, Albert J.R. Heck, Gert Folkers, Lukas C. Kapitein, Simone Lemeer, Paul M.P. van Bergen en Henegouwen and Marc Baldus
This research is conducted within Utrecht University’s research theme Life Sciences and was supported by the Netherlands Organization for Scientific Research (NWO).